Why don't we live on a red planet?

"Why is the grass green?" a classic childhood question, with a classic answer, the chlorophyll pigments in plants reflect mostly green light. But why is the grass really green, and not red or brown? This has been a longstanding question for evolutionary biologists. The quandary is that when photosynthesis entered the eukaryotic domain through primary endosymbiosis, there were actually three players on equal footing: green algae (ultimately giving rise to plants), red algae (their pigment phycoerythrin reflects red light), and the poorly studied glaucophyte algae. So why did the terrestrial environment become green and not red? A bioinformatic study that relied on the largest collection of algal genome data yet assembled, was recently published in the Journal of Phycology by Huan Qiu (a research associate in the Bhattacharya lab) and collaborators that offers a solution to this problem: see also the accompanying piece written by Roscoff scientist and red algal expert, Jonas Collen. It turns out that red algae underwent a drastic phase of gene loss (i.e., known as genome reduction) after its split from green algae (see figure) during which 25% of its core genes were permanently shed. Not only was motility, conferred by the ubiquitous flagella and basal bodies lost but also highly conserved pathways for light sensing (phytochrome), signal transduction and molecular sorting (GPI-anchor biosynthesis; 22 genes), and protein degradation (autophagy; 17 genes). What sort of catastrophe results in such drastic genome streamlining? Qiu et al. posit that the ancestor of red algae survived an extremophilic phase during which they lived in a highly stressful environment (e.g., hot springs such as those found in Yellowstone National Park) that dictated a simplified lifestyle and a tiny genome essential for competing with other small-genomed microbes. Most amazingly, one of these specialized red algal lineages escaped this harsh environment and went on to become a highly successful assemblage of algae and seaweeds living in "normal" habitats such as ponds and the intertidal zone. Qiu et al. suggest that this escape act may have been fueled by horizontal gene transfers from bacteria, that partially compensated for the reduced gene inventory. Regardless, this ancient (> 1 billion years ago) genetic bottleneck likely put red algae at a significant disadvantage in the race for land. The green algae contained ca. 1,700 more genes back then and, as we know, went on to conquer the terrestrial environment. Therefore we are getting closer to understanding why the Earth is a "green" planet, although red and brown algae do quite well in aquatic environments. So, the next time you eat sushi, the wrap is the red alga named Porphyra, or eat ice cream containing carrageenan (extracted from red seaweeds), appreciate the surprising evolutionary path these species took to get to the marketplace, and why we live on a green planet.
Qiu, H., D.C. Price, E.C. Yang, H.S. Yoon, and D. Bhattacharya (2015) Evidence of ancient genome reduction in red algae (Rhodophyta). Journal of Phycology 51:624-636. doi
Collen, J. (2015) Win some, lose some: genome evolution in red algae. Journal of Phycology 51:621-623. doi
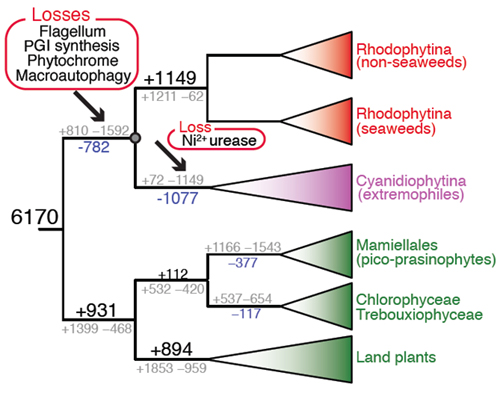
Genome analysis provides estimates of the number of gene family gains and losses (gray numbers at each branch) in the red algae and green algae plus plants. Net gene family gains or losses are shown in black and blue text, respectively. Net gene family changes, when >600 are shown as proportionately sized numbers. The arrows indicate the two major phases of net gene loss in red algae. The major functional losses associated with each phase of genome reduction are also shown.
August 2015


